Our integral genomic signature study for precision oncology published in Nature Communications
May 20, 2022
/
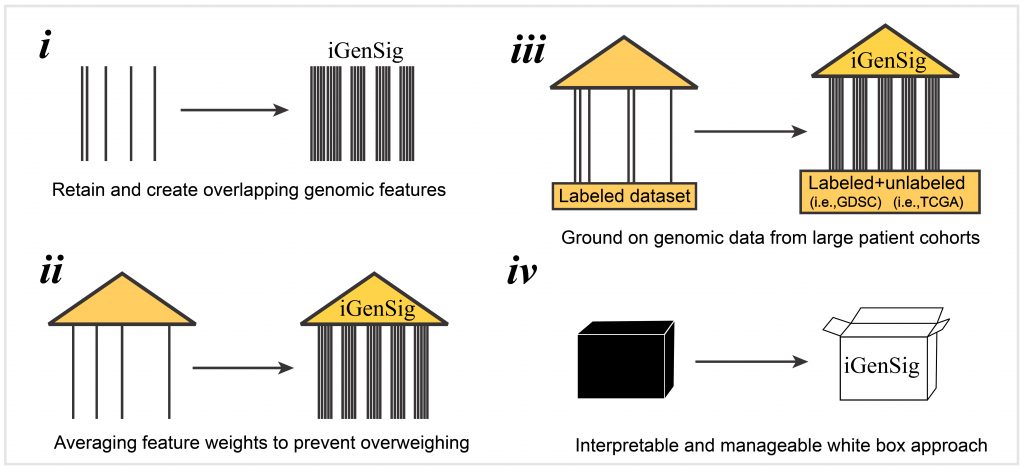
Low-cost multi-omics sequencing is expected to become clinical routine and transform precision oncology. Viable computational methods that can facilitate tailored intervention while tolerating sequencing biases are in high demand. In this study, we develop an integral genomic signature —iGenSig— approach to predict drug responses using multi-omics data from tumour samples, and validate this approach using genomic datasets from six clinical studies and clinical trials. iGenSig will provide a computational framework to empower tailored cancer therapy based on multi-omics data.